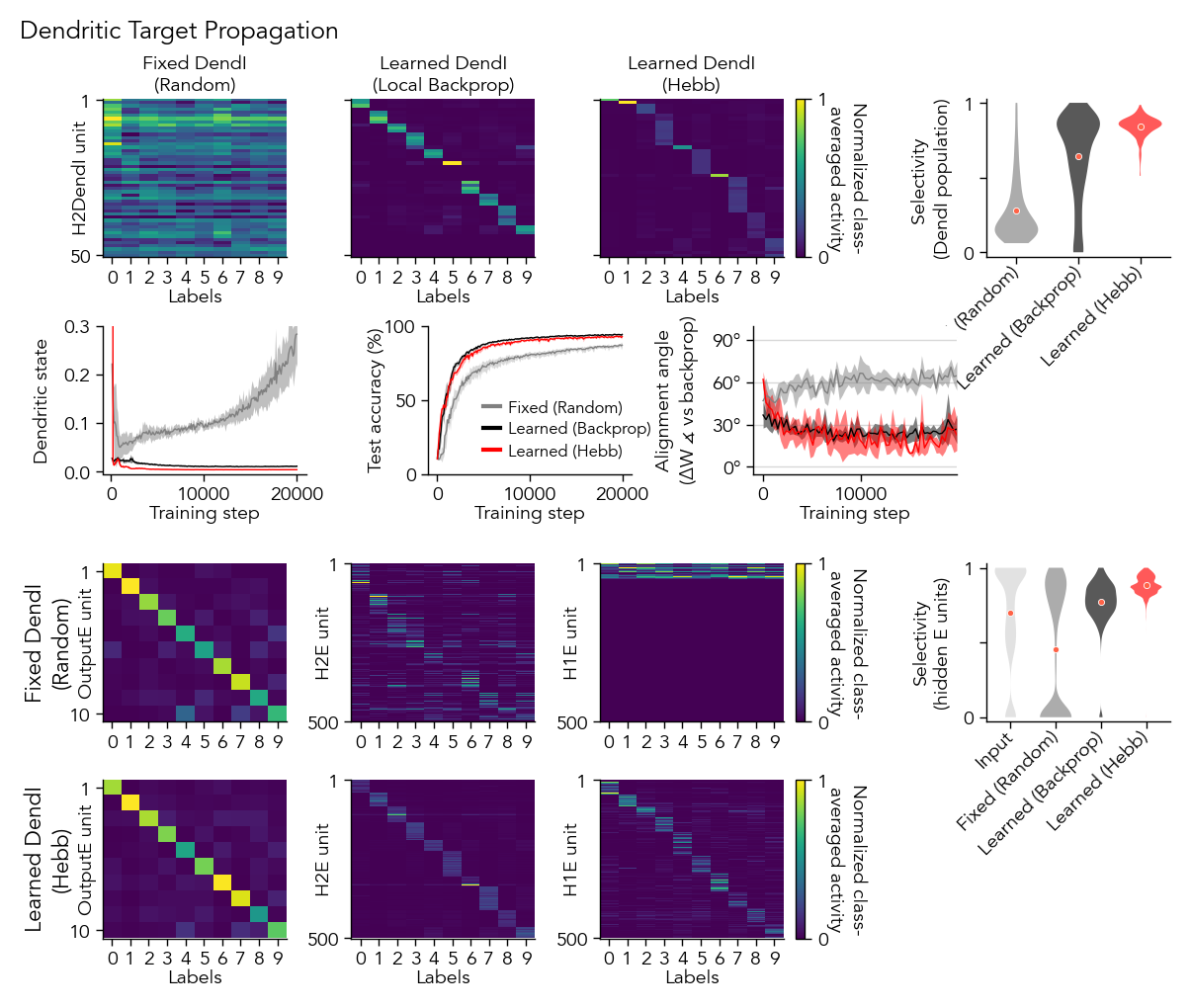
Figure 4: Learning Dend-I weights for Dendritic Target Propagation#
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.gridspec as gs
import h5py
import EIANN as eiann
import EIANN.utils as ut
from EIANN.generate_figures import *
eiann.update_plot_defaults()
figure_name = "Fig4_learning_dendI"
model_list = ["bpLike_WT_fixedDend", "bpLike_WT_localBP", "bpLike_WT_hebbdend"]
model_dict_all = load_model_dict()
generate_hdf5_all_seeds(model_list, model_dict_all, variables_to_save=["average_pop_activity_dict", "metrics_dict", "angle_vs_bp", "dendritic_state"], recompute=None)
Show code cell source
Hide code cell source
fig = plt.figure(figsize=(6, 5))
axes1 = gs.GridSpec(nrows=4, ncols=5, figure=fig,
left=0.08,right=0.98,
bottom=0.07, top=0.92,
wspace=0.4, hspace=0.35,
height_ratios=[1, 1.2, 1, 1], width_ratios=[1, 1, 1, 0.4, 1])
ax_dend_selectivity = fig.add_subplot(axes1[0, 4])
ax_selectivity = fig.add_subplot(axes1[2, 4])
axes2 = gs.GridSpec(nrows=1, ncols=3, figure=fig,
left=0.08,right=0.8,
bottom=0.54, top=0.69,
wspace=0.6, hspace=0.3)
ax_dendstate = fig.add_subplot(axes2[0])
ax_accuracy = fig.add_subplot(axes2[1])
ax_angle = fig.add_subplot(axes2[2])
root_dir = ut.get_project_root()
model_dict_all["bpLike_WT_hebbdend"]["display_name"] = "Learned DendI\n(Hebb)"
model_dict_all["bpLike_WT_hebbdend"]["label"] = "Learned (Hebb)"
model_dict_all["bpLike_WT_localBP"]["display_name"] = "Learned DendI\n(Local Backprop)"
model_dict_all["bpLike_WT_localBP"]["label"] = "Learned (Backprop)"
model_dict_all["bpLike_WT_fixedDend"]["display_name"] = "Fixed DendI\n(Random)"
model_dict_all["bpLike_WT_fixedDend"]["label"] = "Fixed (Random)"
for col, model_key in enumerate(model_list):
model_dict = model_dict_all[model_key]
network_name = model_dict['config'].split('.')[0]
hdf5_path = root_dir + f"/EIANN/data/model_hdf5_plot_data/plot_data_{network_name}.h5"
with h5py.File(hdf5_path, 'r') as f:
data_dict = f[network_name]
# print(f"Generating plots for {model_dict['label']}")
population = 'H2DendI'
example_seed = model_dict['seeds'][0] # example seed to plot
######################################
# Example heatmaps for E populations
######################################
ax = fig.add_subplot(axes1[0, col])
average_pop_activity_dict = data_dict[example_seed]['average_pop_activity_dict']
num_units = average_pop_activity_dict[population].shape[1]
eiann.plot_batch_accuracy_from_data(average_pop_activity_dict, population=population, sort=True, ax=ax, cbar=False)
if col==len(model_list)-1:
pos = ax.get_position()
cbar_ax = fig.add_axes([pos.x1 + 0.01, pos.y0, 0.008, pos.height])
cbar = matplotlib.colorbar.ColorbarBase(cbar_ax, cmap='viridis', orientation='vertical')
cbar.set_label('Normalized class-\naveraged activity', labelpad=14, rotation=270)
cbar.set_ticks([0, 1])
ax.set_yticks([0,num_units-1])
ax.set_yticklabels([1,num_units])
ax.set_ylabel(f'{population} unit', labelpad=-8)
ax.set_xlabel(ax.get_xlabel(), labelpad=0)
ax.set_title(model_dict["display_name"], pad=3)
if col>0:
ax.set_ylabel('')
ax.set_yticklabels([])
######################################
# Model metrics
######################################
plot_accuracy_all_seeds(data_dict, model_dict, ax=ax_accuracy)
legend = ax_accuracy.legend(ncol=1, bbox_to_anchor=(0.2, 0.6), loc='upper left', fontsize=6)
for line in legend.get_lines():
line.set_linewidth(1.5)
plot_dendritic_state_all_seeds(data_dict, model_dict, ax=ax_dendstate)
plot_angle_vs_bp_all_seeds(data_dict, model_dict, ax=ax_angle)
populations_to_plot = [population for population in data_dict[example_seed]['average_pop_activity_dict'] if 'DendI' in population]
plot_metric_all_seeds(data_dict, model_dict, populations_to_plot=populations_to_plot, ax=ax_dend_selectivity, metric_name='selectivity', plot_type='violin', plot_input=False)
ax_dend_selectivity.set_ylabel(f"Selectivity \n(DendI population)")
######################################
# Representations of E populations
######################################
subpanel_config = {"bpLike_WT_fixedDend": {"row": 2, "remove_xlabel": True},
"bpLike_WT_hebbdend": {"row": 3, "remove_xlabel": False}}
if model_key in subpanel_config:
# Plot activity heatmaps of each population to the test dataset
cfg = subpanel_config[model_key]
average_pop_activity_dict = data_dict[example_seed]['average_pop_activity_dict']
for i, population in enumerate(["OutputE", "H2E", "H1E"]):
ax = fig.add_subplot(axes1[cfg["row"], i])
eiann.plot_batch_accuracy_from_data(average_pop_activity_dict, population=population, sort=True, ax=ax, cbar=False)
num_units = average_pop_activity_dict[population].shape[1]
ax.set_yticks([0,num_units-1])
ax.set_yticklabels([1,num_units])
ax.set_ylabel(f'{population} unit', labelpad=-10)
ax.set_xlabel(ax.get_xlabel(), labelpad=0)
if i==0:
ax.set_title(model_dict['display_name'], rotation=90, x=-0.29, y=0.4, va='center', fontsize=8)
if cfg["remove_xlabel"]:
ax.set_xlabel('')
if i==2:
pos = ax.get_position()
cbar_ax = fig.add_axes([pos.x1 + 0.01, pos.y0, 0.008, pos.height])
cbar = matplotlib.colorbar.ColorbarBase(cbar_ax, cmap='viridis', orientation='vertical')
cbar.set_label('Normalized class-\naveraged activity', labelpad=14, rotation=270)
cbar.set_ticks([0, 1])
# Plot sparsity and selectivity of hidden layers
populations_to_plot = ['H1E', 'H2E']
plot_metric_all_seeds(data_dict, model_dict, populations_to_plot=populations_to_plot, ax=ax_selectivity, metric_name='selectivity', plot_type='violin', side='both')
ax_selectivity.set_ylabel(f"Selectivity\n(hidden E units)")
fig.suptitle("Dendritic Target Propagation", x=0.01, y=1., fontsize=9, ha='left')
fig.savefig(f"{root_dir}/EIANN/figures/{figure_name}.png", dpi=300)
fig.savefig(f"{root_dir}/EIANN/figures/{figure_name}.svg", dpi=300)