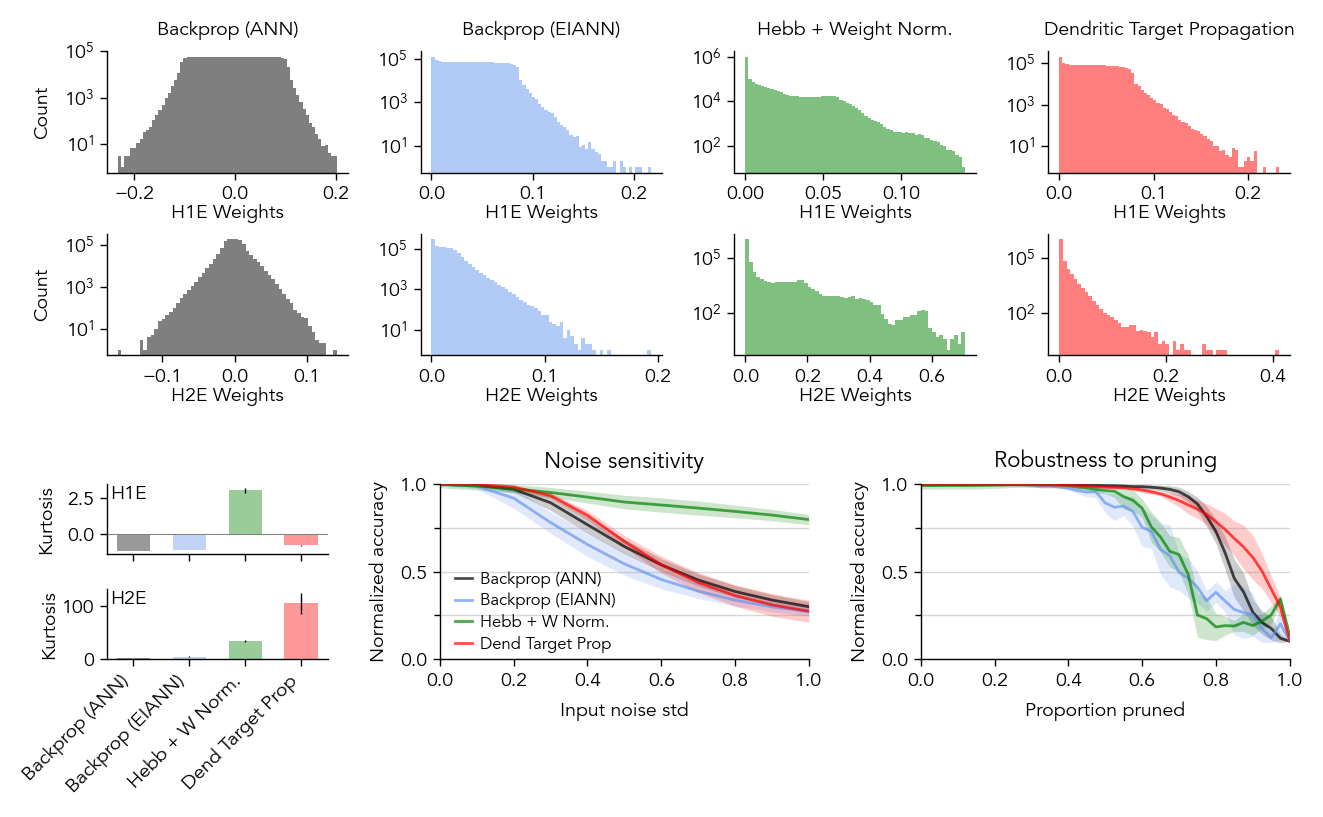
Figure S5: Robustness to perturbations#
import numpy as np
from scipy.stats import kurtosis
import matplotlib.pyplot as plt
import matplotlib.gridspec as gs
import h5py
import EIANN.utils as ut
from EIANN.generate_figures import *
import EIANN as eiann
eiann.update_plot_defaults()
figure_name = "FigS5_robustness_to_perturbations"
model_list = ["vanBP", "bpDale_fixed", "HebbWN_topsup", "bpLike_WT_hebbdend"]
model_dict_all = load_model_dict()
generate_hdf5_all_seeds(model_list, model_dict_all, variables_to_save = ['weights', 'noise_sensitivity', 'robustness_to_pruning'], recompute=None)
Show code cell source
Hide code cell source
fig = plt.figure(figsize=(6.5, 3.8))
axes = gs.GridSpec(nrows=2, ncols=4, figure=fig,
left=0.07,right=0.98,
top=0.92, bottom=0.52,
wspace=0.3, hspace=0.5)
axes_perturbations = gs.GridSpec(nrows=2, ncols=3, figure=fig,
left=0.07,right=0.98,
top=0.35, bottom=0.12,
wspace=0.35, hspace=0.5, width_ratios=[0.6, 1, 1])
ax_kurtosisH1E = fig.add_subplot(axes_perturbations[0, 0])
ax_kurtosisH2E = fig.add_subplot(axes_perturbations[1, 0])
ax_noise = fig.add_subplot(axes_perturbations[:, 1])
ax_pruning = fig.add_subplot(axes_perturbations[:, 2])
root_dir = ut.get_project_root()
model_dict_all["vanBP"]["display_name"] = "Backprop (ANN)"
model_dict_all["bpLike_WT_hebbdend"]["display_name"] = "Dendritic Target Propagation"
model_dict_all["bpDale_fixed"]["display_name"] = "Backprop (EIANN)"
model_dict_all["HebbWN_topsup"]["display_name"] = "Hebb + Weight Norm."
model_dict_all["vanBP"]["label"] = "Backprop (ANN)"
model_dict_all["bpDale_fixed"]["label"] = "Backprop (EIANN)"
model_dict_all["bpLike_WT_hebbdend"]["label"] = "Dend Target Prop"
model_dict_all["HebbWN_topsup"]["label"] = "Hebb + W Norm."
for col, model_key in enumerate(model_list):
model_dict = model_dict_all[model_key]
network_name = model_dict['config'].split('.')[0]
hdf5_path = root_dir + f"/EIANN/data/model_hdf5_plot_data/plot_data_{network_name}.h5"
with h5py.File(hdf5_path, 'r') as f:
data_dict = f[network_name]
# print(f"Generating plots for {model_dict['label']}")
#########################
# Weight distributions
#########################
H1E_weights = []
H2E_weights = []
for seed in model_dict['seeds']:
H1E_weights.extend(data_dict[seed]['weights']['final_weights']['H1E_InputE'][:].flatten())
H2E_weights.extend(data_dict[seed]['weights']['final_weights']['H2E_H1E'][:].flatten())
ax = fig.add_subplot(axes[0, col])
ax.hist(H1E_weights, bins=70, alpha=0.5, color=model_dict["color"], density=False)
ax.set_xlabel("H1E Weights", labelpad=0)
if col == 0:
ax.set_ylabel("Count")
ax.set_title(model_dict['display_name'])
ax.set_yscale('log')
ax = fig.add_subplot(axes[1, col])
ax.hist(H2E_weights, bins=60, alpha=0.5, color=model_dict["color"], density=False)
ax.set_xlabel("H2E Weights", labelpad=0)
if col == 0:
ax.set_ylabel("Count")
ax.set_yscale('log')
projection = 'H1E_InputE'
plot_kurtosis_all_seeds(data_dict, model_dict, projection, ax=ax_kurtosisH1E)
projection = 'H2E_H1E'
plot_kurtosis_all_seeds(data_dict, model_dict, projection, ax=ax_kurtosisH2E)
###############################
# Robustness to input noise
###############################
accuracy_list = []
for seed in model_dict['seeds']:
noise_stds, noise_accuracy = data_dict[seed]['noise_sensitivity']
accuracy_list.append(noise_accuracy)
accuracy_array = np.array(accuracy_list)
mean_accuracy = np.mean(accuracy_array, axis=0)
mean_accuracy_norm = mean_accuracy / np.max(mean_accuracy)
std_accuracy_norm = np.std(accuracy_array, axis=0) / np.max(mean_accuracy)
ax_noise.plot(noise_stds, mean_accuracy_norm, label=model_dict['label'], linewidth=1, color=model_dict["color"], alpha=0.7)
ax_noise.fill_between(noise_stds, mean_accuracy_norm - std_accuracy_norm, mean_accuracy_norm + std_accuracy_norm, alpha=0.2, color=model_dict["color"], linewidth=0.)
ax_noise.set_xlim([0, 1])
ax_noise.set_ylim([0, 1])
ax_noise.set_yticks([0, 0.5, 1], minor=False)
ax_noise.set_yticks(np.linspace(0, 1, 5), minor=True)
ax_noise.legend(frameon=False, fontsize=6, loc='lower left', bbox_to_anchor=(0., -0.05))
ax_noise.grid(True, axis='y', color='gray', linewidth=0.5, alpha=0.3, which='both')
ax_noise.set_xlabel("Input noise std")
ax_noise.set_ylabel("Normalized accuracy")
ax_noise.set_title("Noise sensitivity", fontsize=8)
##################################
# Robustness to synaptic pruning
##################################
accuracy_list = []
for seed in model_dict['seeds']:
fraction_to_prune, accuracy = data_dict[seed]['robustness_to_pruning']
accuracy_list.append(accuracy)
accuracy_array = np.array(accuracy_list)
mean_accuracy = np.mean(accuracy_array, axis=0)
mean_accuracy_norm = mean_accuracy / np.max(mean_accuracy)
std_accuracy_norm = np.std(accuracy_array, axis=0) / np.max(mean_accuracy)
ax_pruning.plot(fraction_to_prune, mean_accuracy_norm, label=model_key, linewidth=1, color=model_dict["color"], alpha=0.7)
ax_pruning.fill_between(fraction_to_prune, mean_accuracy_norm - std_accuracy_norm, mean_accuracy_norm + std_accuracy_norm, alpha=0.2, color=model_dict["color"], linewidth=0.)
ax_pruning.set_xlim([0, 1])
ax_pruning.set_ylim([0, 1])
ax_pruning.set_yticks([0, 0.5, 1], minor=False)
ax_pruning.set_yticks(np.linspace(0, 1, 5), minor=True)
ax_pruning.grid(True, axis='y', color='gray', linewidth=0.5, alpha=0.3, which='both')
ax_pruning.set_xlabel("Proportion pruned")
ax_pruning.set_ylabel("Normalized accuracy")
ax_pruning.set_title("Robustness to pruning", fontsize=8)
ax_kurtosisH1E.set_ylabel("Kurtosis")
ax_kurtosisH2E.set_ylabel("Kurtosis", labelpad=1)
ax_kurtosisH1E.set_title("H1E", x=0.1, y=0.55)
ax_kurtosisH2E.set_title("H2E", x=0.1, y=0.55)
ax_kurtosisH1E.set_xticklabels([])
plt.show()
fig.savefig(f"{root_dir}/EIANN/figures/{figure_name}.svg", dpi=300)
fig.savefig(f"{root_dir}/EIANN/figures/{figure_name}.png", dpi=300)