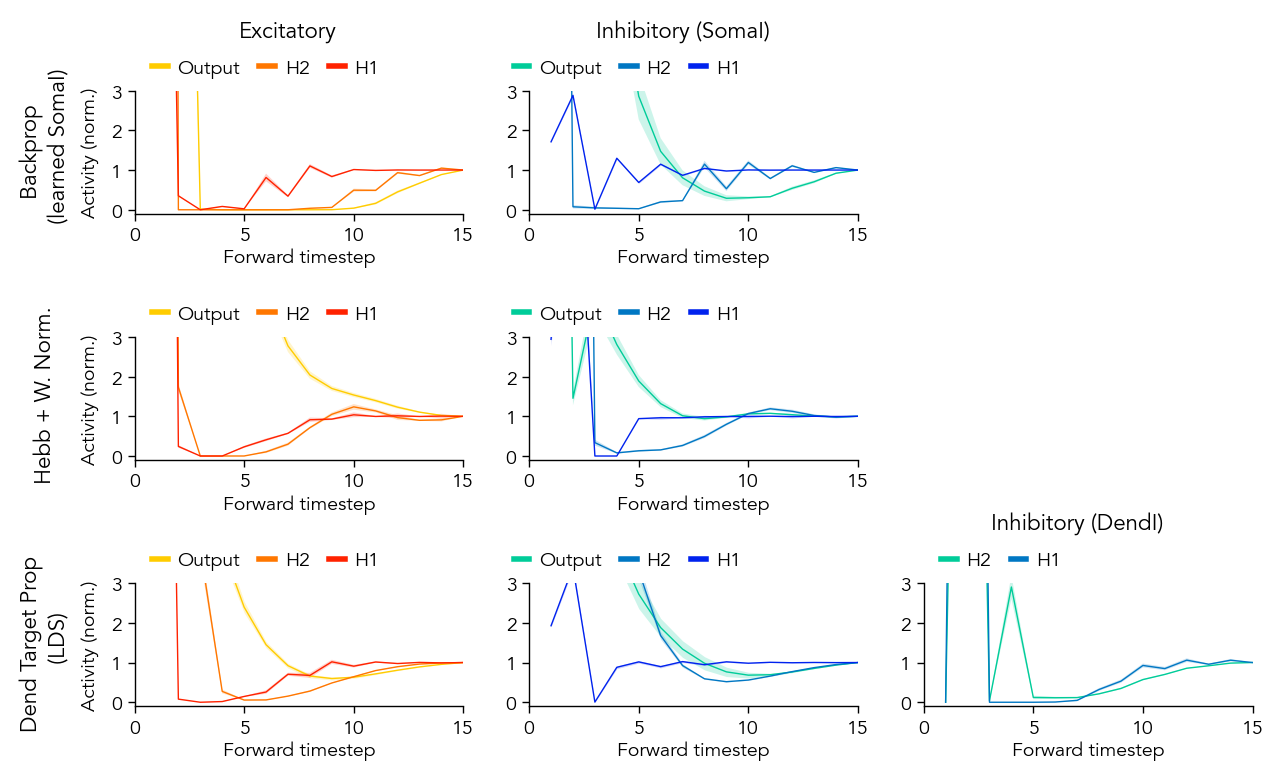
Figure S1: EI dynamics#
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gs
import h5py
import re
import EIANN.utils as ut
from EIANN.generate_figures import *
import EIANN as eiann
eiann.update_plot_defaults()
root_dir = ut.get_project_root()
figure_name = "FigS1_dynamics"
model_list = ["bpDale_learned", "HebbWN_topsup", "bpLike_WT_hebbdend"]
model_dict_all = load_model_dict()
generate_hdf5_all_seeds(model_list, model_dict_all, dataset='MNIST', variables_to_save = ['activity_dynamics'], recompute=None)
Show code cell source
Hide code cell source
fig = plt.figure(figsize=(6.5, 3.8))
axes = gs.GridSpec(nrows=3, ncols=3, figure=fig,
left=0.1,right=0.96,
top=0.89, bottom=0.08,
wspace=0.2, hspace=1)
def get_layer_name(pop_name):
split_strings = re.findall(r'[A-Z][a-z\d]*', pop_name)
return split_strings[0]
model_dict_all["bpDale_learned"]["display_name"] = "Backprop \n (learned SomaI)"
model_dict_all["HebbWN_topsup"]["display_name"] = "Hebb + W. Norm."
model_dict_all["bpLike_WT_hebbdend"]["display_name"] = "Dend Target Prop \n (LDS)"
for row, model_key in enumerate(model_list):
model_dict = model_dict_all[model_key]
network_name = model_dict['config'].split('.')[0]
hdf5_path = root_dir + f"/EIANN/data/model_hdf5_plot_data/plot_data_{network_name}.h5"
with h5py.File(hdf5_path, 'r') as f:
data_dict = f[network_name]
# print(f"Generating plots for {model_dict['label']}")
dynamics_all_seeds = {}
for seed in model_dict['seeds']:
pop_dynamics_dict = data_dict[seed]['activity_dynamics']
for pop_name, pop_dynamics in pop_dynamics_dict.items():
if pop_name not in dynamics_all_seeds:
dynamics_all_seeds[pop_name] = []
avg_dynamics = np.mean(pop_dynamics, axis=(1,2))
avg_dynamics = avg_dynamics/avg_dynamics[-1]
dynamics_all_seeds[pop_name].append(avg_dynamics)
dynamics_all_seeds = {pop_name:np.array(dynamics) for pop_name, dynamics in dynamics_all_seeds.items()}
for col in range(3):
ax = fig.add_subplot(axes[row, col])
if col == 0:
E_populations = {pop_name:pop_dynamics for pop_name,pop_dynamics in dynamics_all_seeds.items() if 'E' in pop_name and 'Input' not in pop_name}
cmap = plt.get_cmap('autumn_r')
for i, (pop_name, pop_dynamics) in enumerate(E_populations.items()):
avg_dynamics = np.mean(pop_dynamics, axis=(0))
error = np.std(pop_dynamics, axis=0)
color = cmap(0.2 + i/3)
ax.plot(np.arange(1,16), avg_dynamics, label=get_layer_name(pop_name), color=color)
ax.fill_between(np.arange(1,16), avg_dynamics-error, avg_dynamics+error, alpha=0.2, linewidth=0, color=color)
legend = ax.legend(ncol=3, loc='upper left', bbox_to_anchor=(-0., 1.4), frameon=False, handlelength=0.8, handletextpad=0.5, columnspacing=1)
for line in legend.get_lines():
line.set_linewidth(2)
elif col == 1:
I_populations = {pop_name:pop_dynamics for pop_name,pop_dynamics in dynamics_all_seeds.items() if 'SomaI' in pop_name}
cmap = plt.get_cmap('winter_r')
for i, (pop_name, pop_dynamics) in enumerate(I_populations.items()):
avg_dynamics = np.mean(pop_dynamics, axis=(0))
error = np.std(pop_dynamics, axis=0)
color = cmap(0.2 + i/3)
ax.plot(np.arange(1,16), avg_dynamics, label=get_layer_name(pop_name), color=color)
ax.fill_between(np.arange(1,16), avg_dynamics-error, avg_dynamics+error, alpha=0.2, linewidth=0, color=color)
legend = ax.legend(ncol=3, loc='upper left', bbox_to_anchor=(-0.1, 1.4), frameon=False, handlelength=0.8, handletextpad=0.5, columnspacing=1)
for line in legend.get_lines():
line.set_linewidth(2)
elif col == 2:
DendI_populations = {pop_name:pop_dynamics for pop_name,pop_dynamics in dynamics_all_seeds.items() if 'DendI' in pop_name}
if len(DendI_populations) > 0:
cmap = plt.get_cmap('winter_r')
for i, (pop_name, pop_dynamics) in enumerate(DendI_populations.items()):
avg_dynamics = np.mean(pop_dynamics, axis=(0))
error = np.std(pop_dynamics, axis=0)
color = cmap(0.2 + i/3)
ax.plot(np.arange(1,16), avg_dynamics, label=get_layer_name(pop_name), color=color)
ax.fill_between(np.arange(1,16), avg_dynamics-error, avg_dynamics+error, alpha=0.2, linewidth=0, color=color)
legend = ax.legend(ncol=3, loc='upper left', bbox_to_anchor=(-0., 1.4), frameon=False, handlelength=0.8, handletextpad=0.5, columnspacing=1)
for line in legend.get_lines():
line.set_linewidth(2)
else:
ax.set_axis_off()
ax.set_xticks(np.arange(0, 16, 5))
ax.set_yticks(np.arange(0, 3.1, 1))
ax.set_ylim([-0.1, 3])
ax.set_xlim([1, 15])
ax.set_xticks([0, 5, 10, 15])
ax.set_xlabel('Forward timestep', labelpad=1)
if col == 0:
ax.set_ylabel('Activity (norm.)', fontsize=7)
ax.set_title(model_dict['display_name'], rotation=90, x=-0.27, y=0.4, va='center', fontsize=8)
if row==0 and col==0:
ax.text(7, 4.5, 'Excitatory', ha='center', va='center', fontsize=8)
elif row==0 and col==1:
ax.text(7, 4.5, 'Inhibitory (SomaI)', ha='center', va='center', fontsize=8)
elif row==2 and col==2:
ax.text(7, 4.5, 'Inhibitory (DendI)', ha='center', va='center', fontsize=8)
fig.savefig(f"{root_dir}/EIANN/figures/{figure_name}.svg", dpi=300)
fig.savefig(f"{root_dir}/EIANN/figures/{figure_name}.png", dpi=300)